RadioGenomic Analyses
- HSIYUN WEI

- Nov 22, 2019
- 1 min read
Updated: Dec 5, 2019
The purpose of this project is to use supervised methods with radiomic, genomic, and survival data. The data file contains a patient number followed by genomic, survival, and radiomic data as follows: The framework of the dataset looks like this:
i) Genomic variables: SMAD4 (0 = false, 1 = true), TP53 (0 = loss of function, 1 = gain of function), and > 4 genes altered (0 = false, 1 = true).
ii) Survival variables: OS_months (number of months since last follow-up or death) and OS_status (0 = alive, 1 = dead).
iii) Radiomic variables: 35 radiomic features extracted from the pancreas tumor in the patient’s CT scan.
Programming Tasks :
Create three models with the classification method of your choice to predict SMAD4, TP53, and number of genes altered (> 4) using the radiomic features. Provide the area under the ROC curve (AUC), sensitivity, and specificity as a table and ROC curve for all models.



2. Investigate the degree of overfitting in task (1) by randomizing the predicted variables (SMAD4, TP53, number of genes altered) and created three new prediction models. Provide the area under the ROC curve (AUC), sensitivity, and specificity as a table and ROC curve for all models.



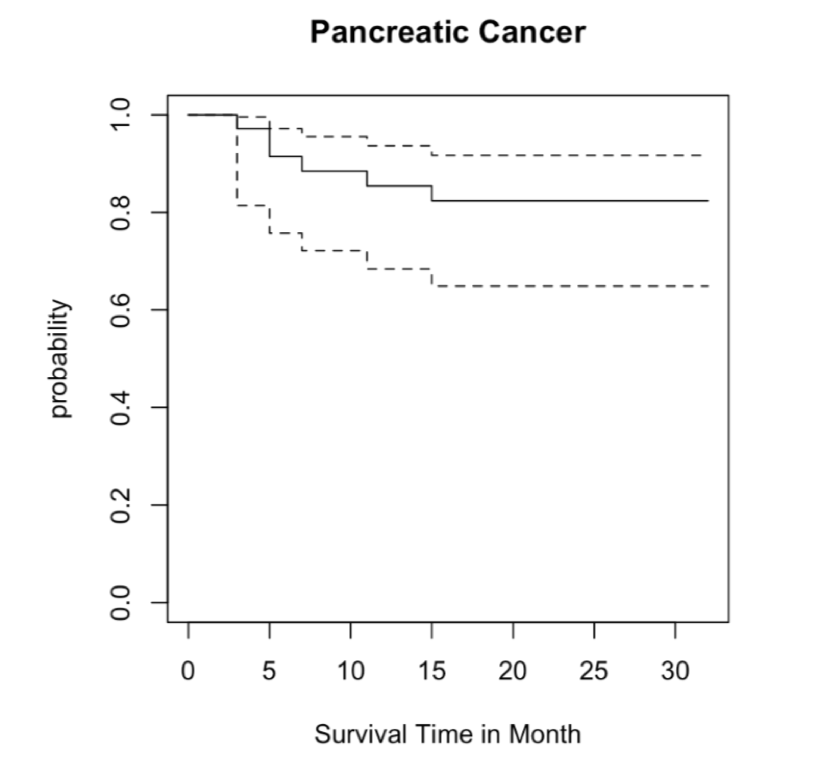

3. Test the association between radiomic and genomic variables with time-dependent survival analysis. Provide a survival curve for the radiomic and genomic variables and a table with p-values.





Comments